New Feature Release: Cell Type Annotations, CNV Inference, Custom Downloads, and More on the ScPCA Portal
Exciting news from the Single-cell Pediatric Cancer Atlas (ScPCA) Portal! All datasets on the portal have been updated to include several new features that enhance data quality and usability. Here’s a close look at what we’ve added and why:
Enhanced Quality Control: Doublet Detection
What’s new:
We performed doublet detection on all samples using [.inline-snippet]scDblFinder[.inline-snippet], a widely used method for identifying doublets in single-cell datasets. The [.inline-snippet]scDblFinder[.inline-snippet] results are now included in both filtered and processed objects, but doublets are not removed.
Why it matters:
By identifying doublets without excluding them, researchers can decide how to handle doublets based on their analysis goals.
Improved Cell Type Annotations
SCimilarity
What’s new:
All samples now include automated cell type annotations from [.inline-snippet]SCimilarity[.inline-snippet], in addition to existing annotations from [.inline-snippet]SingleR[.inline-snippet] and [.inline-snippet]CellAssign[.inline-snippet]. When two out of the three automated methods agree, using an ontology-based approach, a consensus label is assigned.
Why it matters:
We expect this to be most useful for helping researchers quickly identify non-malignant cells, in particular, immune cells. Adding a third automated method enabled us to assign consensus cell types to a larger proportion of cells on the Portal.
Open Single-cell Pediatric Cancer Atlas (OpenScPCA)
What’s new:
As part of the OpenScPCA project, curated cell type annotations have now been generated for two ScPCA datasets:
- Neuroblastoma: [.inline-snippet]SCPCP000004[.inline-snippet]
- Ewing sarcoma: [.inline-snippet]SCPCP000015[.inline-snippet]
Why it matters:
Automated, reference-based methods have considerable limitations when applied to pediatric malignant cells. To tackle this challenge, we launched the OpenScPCA project in 2024 to collaboratively improve cell type annotation.
Learn more in the ScPCA documentation on cell type annotation!
CNV Inference with InferCNV
What’s new:
We performed copy number variation (CNV) inference with [.inline-snippet]InferCNV[.inline-snippet] on samples containing at least 100 non-malignant reference cells, as determined by the updated consensus cell types. The [.inline-snippet]InferCNV[.inline-snippet] output is now available in processed objects.
Why it matters:
CNV inference can provide additional insight by helping to distinguish malignant from non-malignant cells.
Learn more in our documentation on CNV inference. For details on where to find [.inline-snippet]InferCNV[.inline-snippet] results within downloaded objects, see:
Programmatic Downloads with ScPCAr
What’s new:
We’ve created the [.inline-snippet]ScPCAr[.inline-snippet] package, which provides functions to get metadata and download data from the ScPCA Portal programmatically in [.inline-snippet]R[.inline-snippet].
Why it matters:
[.inline-snippet]ScPCAr[.inline-snippet] saves you time by making it easier to work with ScPCA data within [.inline-snippet]R[.inline-snippet].
Explore an example of how to use this package!
Exploratory Visualizations
What’s new:
We launched a new feature, Cell Browser, which enables users to explore interactive visualizations of individual samples directly within the Portal. Our visualization feature is built on UCSC Cell Browser.
Why it matters:
[.inline-snippet]Cell Browser[.inline-snippet] enables researchers to use exploratory, interactive visualizations to gain insights and make informed decisions when working with single-cell transcriptomic data, without first downloading the data.
Create Your Own Custom Downloads
What’s new:
Finally, we’re excited to introduce [.inline-snippet]Datasets[.inline-snippet], a shopping cart-like experience that allows you to build custom downloads. You can combine individual samples and entire projects in a single download.

Now, when you explore an ScPCA project, you’ll see a button that says [.inline-snippet]Add to Dataset[.inline-snippet], which you can select to begin adding that project to your “cart.”
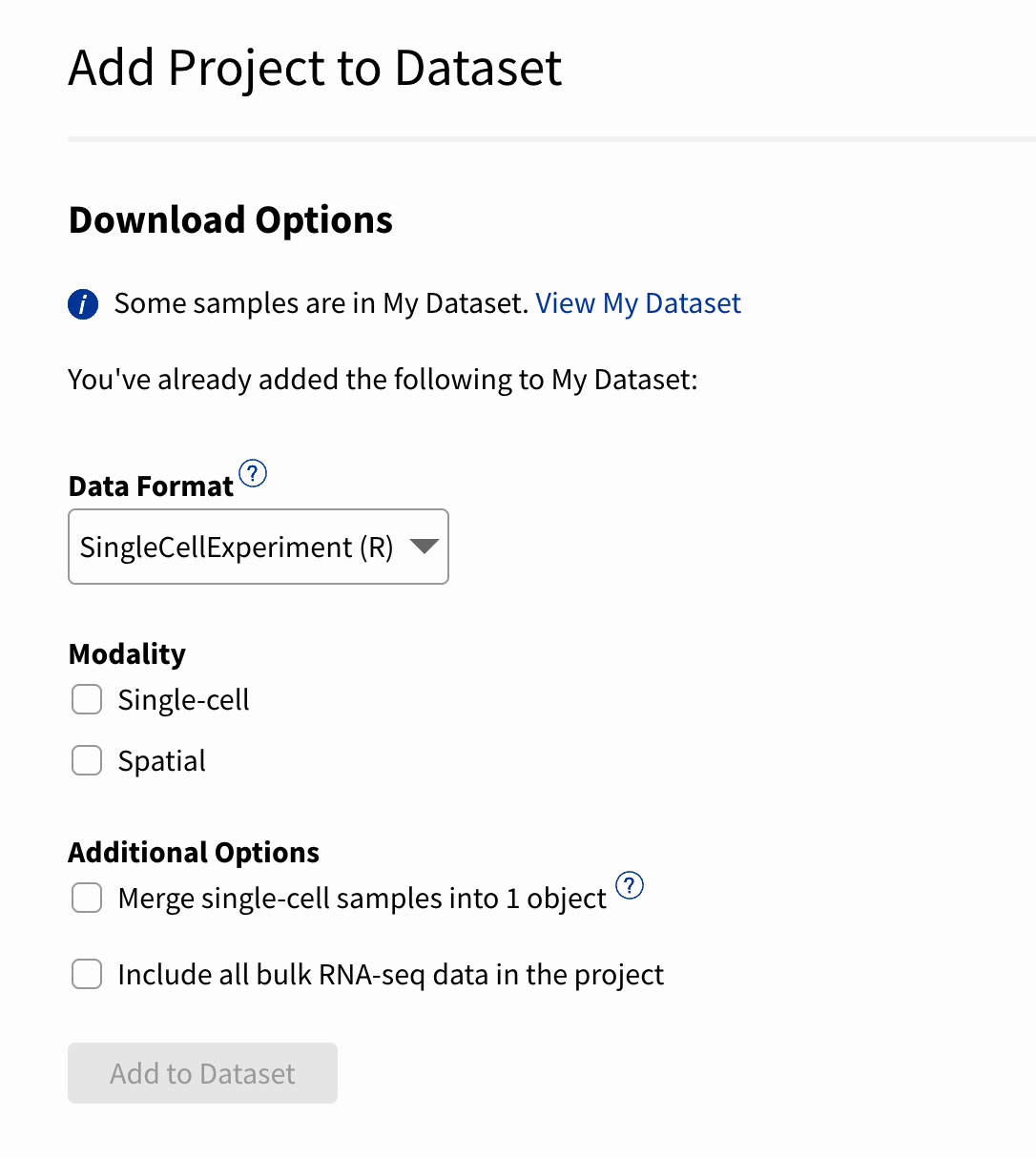
You can then select the data format and any additional modalities (e.g., spatial and bulk) available for the samples in your custom dataset.
Why it matters:
This feature gives you unprecedented control over your ScPCA downloads, helping you tailor your dataset to your own research needs!
Portal-wide Downloads
What’s new:
You can now download all projects from the Portal by data format. Choose to receive your portal-wide download in one of these formats: [.inline-snippet]SingleCellExperiment[.inline-snippet] (R), [.inline-snippet]AnnData[.inline-snippet] (Python), or Spatial (where available). You can still download sample metadata for all projects as well.
Why it matters:
Downloading by format ensures the data is ready for use with your preferred tools. Get started faster with all available data on the Portal!
Get Started Today!
Help Us Test New Features
The Data Lab team is thrilled to share these new features with the community, but we’re not done just yet! We need your help testing the [.inline-snippet]Datasets[.inline-snippet] and [.inline-snippet]Cell Browser[.inline-snippet] features. Your feedback will help us make improvements so we can better support your research and the needs of the broader community. Participants in either study will receive a Data Lab t-shirt as a thank you!
Datasets
If you have downloaded data from the ScPCA portal before or are working with publicly available single-cell pediatric cancer data, we'd love to hear your thoughts about this new feature!
Sign up to spend one hour with our UXR Designer to explore [.inline-snippet]Datasets[.inline-snippet], while telling us about your experience.
Cell Browser
We’re also conducting a study to understand how you use exploratory visualizations to make decisions while working with single-cell transcriptomic data.
Are you a researcher who has used the ScPCA Portal before? Are you new to using [.inline-snippet]R[.inline-snippet] to visualize data? If so, we hope you’ll join our UXR designer to try this new feature and share your thoughts during a one hour virtual session.
These updates were built with researchers like you in mind and we can’t wait for you to try them out! Have questions or suggestions? Reach out to us at scpca@alexslemonade.org.
Exciting news from the Single-cell Pediatric Cancer Atlas (ScPCA) Portal! All datasets on the portal have been updated to include several new features that enhance data quality and usability. Here’s a close look at what we’ve added and why:
Enhanced Quality Control: Doublet Detection
What’s new:
We performed doublet detection on all samples using [.inline-snippet]scDblFinder[.inline-snippet], a widely used method for identifying doublets in single-cell datasets. The [.inline-snippet]scDblFinder[.inline-snippet] results are now included in both filtered and processed objects, but doublets are not removed.
Why it matters:
By identifying doublets without excluding them, researchers can decide how to handle doublets based on their analysis goals.
Improved Cell Type Annotations
SCimilarity
What’s new:
All samples now include automated cell type annotations from [.inline-snippet]SCimilarity[.inline-snippet], in addition to existing annotations from [.inline-snippet]SingleR[.inline-snippet] and [.inline-snippet]CellAssign[.inline-snippet]. When two out of the three automated methods agree, using an ontology-based approach, a consensus label is assigned.
Why it matters:
We expect this to be most useful for helping researchers quickly identify non-malignant cells, in particular, immune cells. Adding a third automated method enabled us to assign consensus cell types to a larger proportion of cells on the Portal.
Open Single-cell Pediatric Cancer Atlas (OpenScPCA)
What’s new:
As part of the OpenScPCA project, curated cell type annotations have now been generated for two ScPCA datasets:
- Neuroblastoma: [.inline-snippet]SCPCP000004[.inline-snippet]
- Ewing sarcoma: [.inline-snippet]SCPCP000015[.inline-snippet]
Why it matters:
Automated, reference-based methods have considerable limitations when applied to pediatric malignant cells. To tackle this challenge, we launched the OpenScPCA project in 2024 to collaboratively improve cell type annotation.
Learn more in the ScPCA documentation on cell type annotation!
CNV Inference with InferCNV
What’s new:
We performed copy number variation (CNV) inference with [.inline-snippet]InferCNV[.inline-snippet] on samples containing at least 100 non-malignant reference cells, as determined by the updated consensus cell types. The [.inline-snippet]InferCNV[.inline-snippet] output is now available in processed objects.
Why it matters:
CNV inference can provide additional insight by helping to distinguish malignant from non-malignant cells.
Learn more in our documentation on CNV inference. For details on where to find [.inline-snippet]InferCNV[.inline-snippet] results within downloaded objects, see:
Programmatic Downloads with ScPCAr
What’s new:
We’ve created the [.inline-snippet]ScPCAr[.inline-snippet] package, which provides functions to get metadata and download data from the ScPCA Portal programmatically in [.inline-snippet]R[.inline-snippet].
Why it matters:
[.inline-snippet]ScPCAr[.inline-snippet] saves you time by making it easier to work with ScPCA data within [.inline-snippet]R[.inline-snippet].
Explore an example of how to use this package!
Exploratory Visualizations
What’s new:
We launched a new feature, Cell Browser, which enables users to explore interactive visualizations of individual samples directly within the Portal. Our visualization feature is built on UCSC Cell Browser.
Why it matters:
[.inline-snippet]Cell Browser[.inline-snippet] enables researchers to use exploratory, interactive visualizations to gain insights and make informed decisions when working with single-cell transcriptomic data, without first downloading the data.
Create Your Own Custom Downloads
What’s new:
Finally, we’re excited to introduce [.inline-snippet]Datasets[.inline-snippet], a shopping cart-like experience that allows you to build custom downloads. You can combine individual samples and entire projects in a single download.

Now, when you explore an ScPCA project, you’ll see a button that says [.inline-snippet]Add to Dataset[.inline-snippet], which you can select to begin adding that project to your “cart.”

You can then select the data format and any additional modalities (e.g., spatial and bulk) available for the samples in your custom dataset.
Why it matters:
This feature gives you unprecedented control over your ScPCA downloads, helping you tailor your dataset to your own research needs!
Portal-wide Downloads
What’s new:
You can now download all projects from the Portal by data format. Choose to receive your portal-wide download in one of these formats: [.inline-snippet]SingleCellExperiment[.inline-snippet] (R), [.inline-snippet]AnnData[.inline-snippet] (Python), or Spatial (where available). You can still download sample metadata for all projects as well.
Why it matters:
Downloading by format ensures the data is ready for use with your preferred tools. Get started faster with all available data on the Portal!
Get Started Today!
Help Us Test New Features
The Data Lab team is thrilled to share these new features with the community, but we’re not done just yet! We need your help testing the [.inline-snippet]Datasets[.inline-snippet] and [.inline-snippet]Cell Browser[.inline-snippet] features. Your feedback will help us make improvements so we can better support your research and the needs of the broader community. Participants in either study will receive a Data Lab t-shirt as a thank you!
Datasets
If you have downloaded data from the ScPCA portal before or are working with publicly available single-cell pediatric cancer data, we'd love to hear your thoughts about this new feature!
Sign up to spend one hour with our UXR Designer to explore [.inline-snippet]Datasets[.inline-snippet], while telling us about your experience.
Cell Browser
We’re also conducting a study to understand how you use exploratory visualizations to make decisions while working with single-cell transcriptomic data.
Are you a researcher who has used the ScPCA Portal before? Are you new to using [.inline-snippet]R[.inline-snippet] to visualize data? If so, we hope you’ll join our UXR designer to try this new feature and share your thoughts during a one hour virtual session.
These updates were built with researchers like you in mind and we can’t wait for you to try them out! Have questions or suggestions? Reach out to us at scpca@alexslemonade.org.

