Introducing the ScPCA downstream analysis workflow!
At the Data Lab, we are constantly looking for ways to enhance the tools we build for pediatric cancer researchers. Earlier this year, we launched the Single-cell Pediatric Cancer Atlas portal, a database of uniformly-processed single-cell data from pediatric cancer clinical samples. One way we felt the portal could be even more beneficial to pediatric cancer researchers is with a ready-to-go workflow that takes in single-cell data and prepares it for downstream analyses such as unsupervised clustering.
We put together the ScPCA downstream analysis workflow to filter, normalize, and perform dimensionality reduction, as well as incorporate initial clustering results to each processed sample/library object.
What the workflow does
The workflow takes in pre-processed gene expression data stored as a SingleCellExperiment object (filtered to remove empty cells/droplets), and performs the following steps:
- Filtering - Each library is filtered to remove any low quality cells using either miQC::filterCells() or through setting a series of manual thresholds (e.g. the minimum number of UMI counts). Genes found in a low percentage of cells in each library are also removed at this step.
- Normalization - Cells are normalized using the deconvolution method from Lun, Bach, and Marioni (2016). The normalized log counts are stored in the SingleCellExperiment object returned by the workflow.
- Dimensionality Reduction - Reduced dimensions are calculated using both principal component analysis (PCA) and uniform manifold approximation and projection (UMAP). The embeddings from PCA and UMAP can be found in the SingleCellExperiment object returned by the workflow.
- Clustering - Cells are assigned to cell clusters using graph-based clustering. By default, louvain clustering is performed using the bluster::NNGraphParam() function with a default nearest-neighbors parameter of 10. Alternatively, walktrap graph-based clustering can be specified, and the number of nearest neighbors parameter can be altered if desired. Cluster assignments are stored in the SingleCellExperiment object returned by the workflow.
What the workflow does not do
The workflow is meant to be a user-friendly, ready-to-go workflow that will perform the core steps often needed when analyzing single-cell data. However, there are a few things that the workflow will not do:
- The workflow will not pre-process your data. Instead, you can see our scpca-nf workflow and ScPCA Portal docs for our suggested approach to pre-processing your data before implementing this workflow, which includes filtering to remove empty cells/droplets.
- The workflow will not convert your data into a SingleCellExperiment object, which means that if your data exists in other formats (e.g. a Seurat object), you will need to perform the initial conversion yourself.
- The workflow will not make decisions about your data. You will be provided with results based on the parameters used throughout the workflow, but we do not do the science for you! You will need to look at the results for yourself to interpret whether or not they are reasonable. Decision making can be highly dependent on the needs of individual researchers, so making such decisions for everyone would not be possible or beneficial.
Workflow requirements
To run the downstream analyses workflow on your own sample data, you will need the following:
- Single-cell gene expression data stored as SingleCellExperiment objects in RDS files. The workflow is set up to process data available on the Single-cell Pediatric Cancer Atlas portal and output from the scpca-nf workflow where single-cell/single-nuclei gene expression data is mapped and quantified using alevin-fry. For more information on this pre-processing, please see the ScPCA Portal docs. Note however that the input for this pipeline is not required to be scpca-nf processed output; the only requirement is that gene expression data be stored as a SingleCellExperiment object in an RDS file.
- A project metadata tab-separated value (TSV) file containing relevant information about the sample data necessary for processing, as described in the metadata file format section of the GitHub repository’s README file.
- A mitochondrial gene list that is specific to the genome or transcriptome version used for alignment. The workflow will use the mitochondrial gene list obtained from Ensembl version 104 by default. This gene list can be found in the reference-files directory of the Github repository.
- A local installation of Snakemake and either R or conda. (See more on this in the how to install the core downstream analyses workflow section.)
Expected output
For each SingleCellExperiment associated with an individual library, the workflow will return two files: a processed SingleCellExperiment object stored in an RDS file containing normalized data and clustering results, and a summary HTML report detailing the filtering of low-quality cells, dimensionality reduction, and clustering that was performed within the workflow. Below is an example of the expected output file structure.
The first output file is the *<library_id>_<filtering_method>_processed_sce.rds file, where `library_id` is the unique library and `filtering_method` is one of miQC or manual. This is the RDS file that contains the final processed SingleCellExperiment object, which contains the filtered, normalized data and clustering results. Clustering results can be found in the colData of the SingleCellExperiment object, stored in a metadata column named using the associated clustering type and nearest neighbors values.
The *<library_id>_<filtering_method>_core_analysis_report.html file is the html file that contains the summary report of the filtering, dimensionality reduction, and clustering results associated with the processed SingleCellExperiment object.
Walk through an example with us!
After ensuring that all of the required software is installed on your machine, as described in the how to install the core downstream analyses workflow section of the GitHub repository’s README file, we are ready to get started with the workflow!
Step 1 - Set up input data
Once you confirm that your single-cell gene expression data has been stored as SingleCellExperiment objects within RDS files, where each RDS file is associated with a single library id, you are ready to set up the project metadata file.
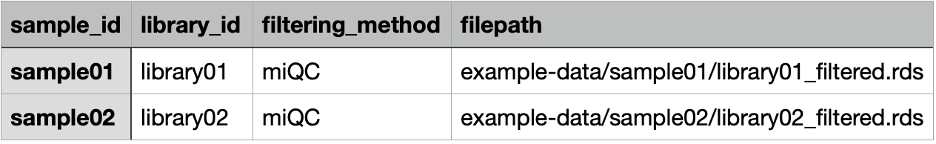
Create a new TSV file, where each row corresponds to a unique library and the columns contain the information associated with that library, and save it in the project-metadata directory. Below is an example of the columns and values that should be in the project metadata TSV file. See more on the columns and their expected values at the metadata file format section of the Github repository’s README.
Step 2 - Run the workflow via command line
Now we can run the snakemake workflow via the command line. To run the workflow we will also need to specify a few parameters, such as where to store the output, the path to the project metadata file, and the path to the mitochondrial gene list. We specify this information using the--config flag which allows us to override their default values for the relevant variables found in the config.yaml file. Below is an example of running the workflow via the command line while modifying only the necessary config file variables.
Step 3 - Check output!
Upon successful completion of the workflow, you should have two output files per sample/library id specified in the project metadata file. Below is the output of the test sample data we ran.
How to get started!
If you are interested in trying the ScPCA downstream analysis workflow on your data or are just curious to learn a bit more about how it works, you can get started with the README associated with the GitHub repository! We also have provided documentation with further details on the processing steps and decisions made throughout the workflow.
Additionally, if you are interested in participating in usability testing of the downstream analysis workflow, please fill out this form and we will contact you when we are ready for testing!
If you have questions about the ScPCA portal, you can reach out to us at scpca@ccdatalab.org!
At the Data Lab, we are constantly looking for ways to enhance the tools we build for pediatric cancer researchers. Earlier this year, we launched the Single-cell Pediatric Cancer Atlas portal, a database of uniformly-processed single-cell data from pediatric cancer clinical samples. One way we felt the portal could be even more beneficial to pediatric cancer researchers is with a ready-to-go workflow that takes in single-cell data and prepares it for downstream analyses such as unsupervised clustering.
We put together the ScPCA downstream analysis workflow to filter, normalize, and perform dimensionality reduction, as well as incorporate initial clustering results to each processed sample/library object.
What the workflow does
The workflow takes in pre-processed gene expression data stored as a SingleCellExperiment object (filtered to remove empty cells/droplets), and performs the following steps:
- Filtering - Each library is filtered to remove any low quality cells using either miQC::filterCells() or through setting a series of manual thresholds (e.g. the minimum number of UMI counts). Genes found in a low percentage of cells in each library are also removed at this step.
- Normalization - Cells are normalized using the deconvolution method from Lun, Bach, and Marioni (2016). The normalized log counts are stored in the SingleCellExperiment object returned by the workflow.
- Dimensionality Reduction - Reduced dimensions are calculated using both principal component analysis (PCA) and uniform manifold approximation and projection (UMAP). The embeddings from PCA and UMAP can be found in the SingleCellExperiment object returned by the workflow.
- Clustering - Cells are assigned to cell clusters using graph-based clustering. By default, louvain clustering is performed using the bluster::NNGraphParam() function with a default nearest-neighbors parameter of 10. Alternatively, walktrap graph-based clustering can be specified, and the number of nearest neighbors parameter can be altered if desired. Cluster assignments are stored in the SingleCellExperiment object returned by the workflow.
What the workflow does not do
The workflow is meant to be a user-friendly, ready-to-go workflow that will perform the core steps often needed when analyzing single-cell data. However, there are a few things that the workflow will not do:
- The workflow will not pre-process your data. Instead, you can see our scpca-nf workflow and ScPCA Portal docs for our suggested approach to pre-processing your data before implementing this workflow, which includes filtering to remove empty cells/droplets.
- The workflow will not convert your data into a SingleCellExperiment object, which means that if your data exists in other formats (e.g. a Seurat object), you will need to perform the initial conversion yourself.
- The workflow will not make decisions about your data. You will be provided with results based on the parameters used throughout the workflow, but we do not do the science for you! You will need to look at the results for yourself to interpret whether or not they are reasonable. Decision making can be highly dependent on the needs of individual researchers, so making such decisions for everyone would not be possible or beneficial.
Workflow requirements
To run the downstream analyses workflow on your own sample data, you will need the following:
- Single-cell gene expression data stored as SingleCellExperiment objects in RDS files. The workflow is set up to process data available on the Single-cell Pediatric Cancer Atlas portal and output from the scpca-nf workflow where single-cell/single-nuclei gene expression data is mapped and quantified using alevin-fry. For more information on this pre-processing, please see the ScPCA Portal docs. Note however that the input for this pipeline is not required to be scpca-nf processed output; the only requirement is that gene expression data be stored as a SingleCellExperiment object in an RDS file.
- A project metadata tab-separated value (TSV) file containing relevant information about the sample data necessary for processing, as described in the metadata file format section of the GitHub repository’s README file.
- A mitochondrial gene list that is specific to the genome or transcriptome version used for alignment. The workflow will use the mitochondrial gene list obtained from Ensembl version 104 by default. This gene list can be found in the reference-files directory of the Github repository.
- A local installation of Snakemake and either R or conda. (See more on this in the how to install the core downstream analyses workflow section.)
Expected output
For each SingleCellExperiment associated with an individual library, the workflow will return two files: a processed SingleCellExperiment object stored in an RDS file containing normalized data and clustering results, and a summary HTML report detailing the filtering of low-quality cells, dimensionality reduction, and clustering that was performed within the workflow. Below is an example of the expected output file structure.
The first output file is the *<library_id>_<filtering_method>_processed_sce.rds file, where `library_id` is the unique library and `filtering_method` is one of miQC or manual. This is the RDS file that contains the final processed SingleCellExperiment object, which contains the filtered, normalized data and clustering results. Clustering results can be found in the colData of the SingleCellExperiment object, stored in a metadata column named using the associated clustering type and nearest neighbors values.
The *<library_id>_<filtering_method>_core_analysis_report.html file is the html file that contains the summary report of the filtering, dimensionality reduction, and clustering results associated with the processed SingleCellExperiment object.
Walk through an example with us!
After ensuring that all of the required software is installed on your machine, as described in the how to install the core downstream analyses workflow section of the GitHub repository’s README file, we are ready to get started with the workflow!
Step 1 - Set up input data
Once you confirm that your single-cell gene expression data has been stored as SingleCellExperiment objects within RDS files, where each RDS file is associated with a single library id, you are ready to set up the project metadata file.
Create a new TSV file, where each row corresponds to a unique library and the columns contain the information associated with that library, and save it in the project-metadata directory. Below is an example of the columns and values that should be in the project metadata TSV file. See more on the columns and their expected values at the metadata file format section of the Github repository’s README.

Step 2 - Run the workflow via command line
Now we can run the snakemake workflow via the command line. To run the workflow we will also need to specify a few parameters, such as where to store the output, the path to the project metadata file, and the path to the mitochondrial gene list. We specify this information using the--config flag which allows us to override their default values for the relevant variables found in the config.yaml file. Below is an example of running the workflow via the command line while modifying only the necessary config file variables.
Step 3 - Check output!
Upon successful completion of the workflow, you should have two output files per sample/library id specified in the project metadata file. Below is the output of the test sample data we ran.
How to get started!
If you are interested in trying the ScPCA downstream analysis workflow on your data or are just curious to learn a bit more about how it works, you can get started with the README associated with the GitHub repository! We also have provided documentation with further details on the processing steps and decisions made throughout the workflow.
Additionally, if you are interested in participating in usability testing of the downstream analysis workflow, please fill out this form and we will contact you when we are ready for testing!
If you have questions about the ScPCA portal, you can reach out to us at scpca@ccdatalab.org!

