Does Bulk Tissue Still Belong in a Single-Cell Atlas?
Earlier this year, Alex’s Lemonade Stand Foundation identified single-cell gene expression profiling as an opportunity to build an atlas of cell types within tumors that could be broadly reused by pediatric cancer researchers. We in the Childhood Cancer Data Lab worked with the grant-making side of ALSF to design a Single-cell Pediatric Cancer Atlas RFA that would maximize the reusability of atlas datasets. One of the bullet points in the RFA that we’ve gotten a number of questions about is “applicants are required to perform RNA-sequencing of the bulk tissue from which the single cells are derived.” We wanted to take this opportunity to outline why we thought that including bulk sequencing would maximize reusability. N.B.: This post does not contain all available references on a topic: we selected a few from many possible references. Commenting is enabled on this post for the next month, so feel free to leave your favorite references on the topic as well.
Let’s briefly define what we mean by single-cell and bulk profiling. When we refer to bulk gene expression profiling, we’re discussing a process whereby RNA from throughout that tissue is profiled together. Bulk profiling can’t easily tell us what’s going on within each individual cell, but we do get to observe the extent to which each gene is expressed in that section of tissue. Single-cell profiling involves a step where bulk tissue is separated into individual cells, and then gene expression levels in those cells are measured separately for each cell.
Why build a single-cell pediatric cancer atlas?
While pediatric cancers are, collectively, the largest killer by disease of children in the US, they are each individually rare. This means that we couldn’t count on a future where each scientist would be able to rapidly build their own single-cell dataset from fresh tissues for each cancer type of interest. We needed a design that would allow researchers who have existing datasets, which were profiled from bulk tissue, to better understand their own data in the context of the single-cell atlas projects.
The arguments against requiring bulk profiling
- The coexpression levels of genes in bulk samples, especially of non-malignant tissue, is often driven by differences in cell-type composition. [Farahbod & Pavlidis]
- It’s possible to create pseudo-bulk data by aggregating data from single cells. [Ding et al. Sup Fig 10]
- There are deconvolution strategies that can use expression profiles from single-cell RNA-seq data to estimate the relative proportions of cell types in bulk-profiled samples. [Jew et al. among a growing number of others]
The arguments for requiring bulk profiling
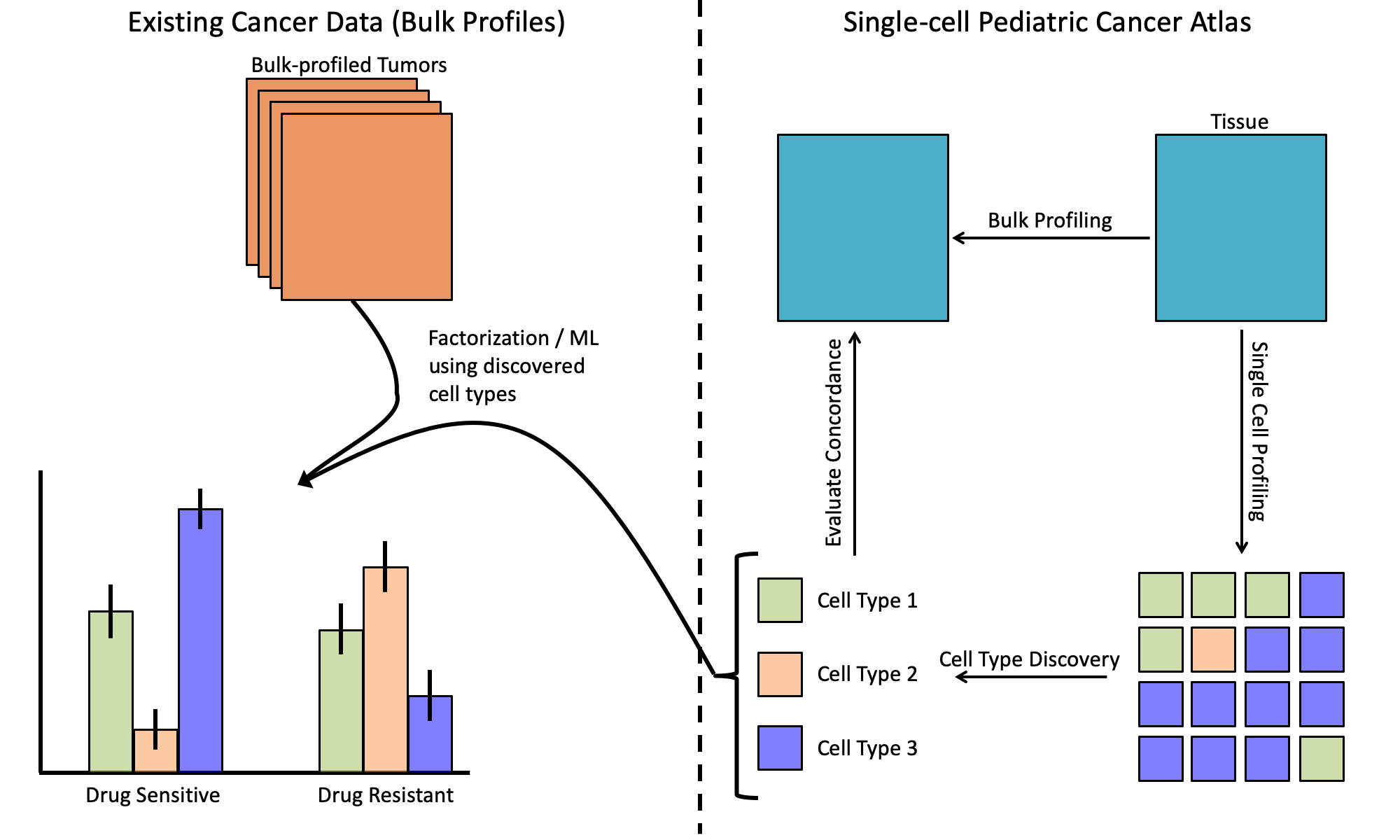
- Dissociation is a physical process. Even if the pseudo-bulk profiles for certain tissue types display a high concordance with bulk, there is no guarantee that this concordance will still be high in these malignant tissues. For example, the data from Ding et al. focuses on normal tissue. If certain cell types are preferentially lost during dissociation then pseudo-bulk would be biased in different ways from traditional bulk measurement, which could make comparison difficult. Imagine that in the figure above, Cell Type 1 is lost during single-cell profiling. There is some risk that the atlas could fail to identify one or more cell types.
- Deconvolution strategies could still be helpful as long as cell-type reference profiles could be defined. However, this requires that the profiles of each cell type be available. Though this appears to often be the case for normal tissue. However, Suvà and Tirosh note in their recent review, “An emerging theme from recent scRNA-seq studies is that malignant cells tend to cluster in their expression profiles primarily by patient sample, and non-malignant cells cluster in their expression profiles by cell type, somewhat independently of the patient of origin (Filbin et al., 2018; Jerby-Arnon et al.,2018; Tirosh et al., 2016a, 2016b; Venteicher et al., 2017). This indicates that (1) inter-tumor heterogeneity is typically larger for malignant cells than for any particular type of non-malignant cells. (2) For malignant cells, inter-tumor heterogeneity is much larger than intra-tumor heterogeneity.” Imagine in the figure above that Cell Type 2’s expression differs greatly from patient-to-patient. There is some risk that deconvolution may be less successful if the inter-tumor expression heterogeneity for malignant cells is large for one or more of the profiled cancer types.
Why we asked for bulk profiling
On balance, there were enough unknown factors or areas of potential risk with a single-cell only approach for malignant tissue profiling that we elected to require bulk sequencing. As the RFA was being designed, it was not clear what the right approach would be for pediatric tumor tissues. These cancers are often somewhat biologically distinct from the adult cancers that much of the literature-to-date for single-cell profiling of human tumors has focused on. There have been a fair number of single-cell profiles of both PDX and mouse model samples, but it wasn’t clear if that data would be directly applicable to an atlas of human tumors.
Looking at the available literature and the uncertainties that existed, we thought it would be prudent to request candidates obtain bulk assays at the same time given the relative risks and benefits plus the modest cost of bulk sequencing relative to single-cell profiling. This means that we’ll be able to evaluate the concordance between single-cell profiles and what is observed in bulk samples. We hope that the references and literature can help others who are independently designing studies of malignancies, particularly pediatric ones, to best allocate resources towards their own efforts.
Earlier this year, Alex’s Lemonade Stand Foundation identified single-cell gene expression profiling as an opportunity to build an atlas of cell types within tumors that could be broadly reused by pediatric cancer researchers. We in the Childhood Cancer Data Lab worked with the grant-making side of ALSF to design a Single-cell Pediatric Cancer Atlas RFA that would maximize the reusability of atlas datasets. One of the bullet points in the RFA that we’ve gotten a number of questions about is “applicants are required to perform RNA-sequencing of the bulk tissue from which the single cells are derived.” We wanted to take this opportunity to outline why we thought that including bulk sequencing would maximize reusability. N.B.: This post does not contain all available references on a topic: we selected a few from many possible references. Commenting is enabled on this post for the next month, so feel free to leave your favorite references on the topic as well.
Let’s briefly define what we mean by single-cell and bulk profiling. When we refer to bulk gene expression profiling, we’re discussing a process whereby RNA from throughout that tissue is profiled together. Bulk profiling can’t easily tell us what’s going on within each individual cell, but we do get to observe the extent to which each gene is expressed in that section of tissue. Single-cell profiling involves a step where bulk tissue is separated into individual cells, and then gene expression levels in those cells are measured separately for each cell.
Why build a single-cell pediatric cancer atlas?
While pediatric cancers are, collectively, the largest killer by disease of children in the US, they are each individually rare. This means that we couldn’t count on a future where each scientist would be able to rapidly build their own single-cell dataset from fresh tissues for each cancer type of interest. We needed a design that would allow researchers who have existing datasets, which were profiled from bulk tissue, to better understand their own data in the context of the single-cell atlas projects.

The arguments against requiring bulk profiling
- The coexpression levels of genes in bulk samples, especially of non-malignant tissue, is often driven by differences in cell-type composition. [Farahbod & Pavlidis]
- It’s possible to create pseudo-bulk data by aggregating data from single cells. [Ding et al. Sup Fig 10]
- There are deconvolution strategies that can use expression profiles from single-cell RNA-seq data to estimate the relative proportions of cell types in bulk-profiled samples. [Jew et al. among a growing number of others]
The arguments for requiring bulk profiling
- Dissociation is a physical process. Even if the pseudo-bulk profiles for certain tissue types display a high concordance with bulk, there is no guarantee that this concordance will still be high in these malignant tissues. For example, the data from Ding et al. focuses on normal tissue. If certain cell types are preferentially lost during dissociation then pseudo-bulk would be biased in different ways from traditional bulk measurement, which could make comparison difficult. Imagine that in the figure above, Cell Type 1 is lost during single-cell profiling. There is some risk that the atlas could fail to identify one or more cell types.
- Deconvolution strategies could still be helpful as long as cell-type reference profiles could be defined. However, this requires that the profiles of each cell type be available. Though this appears to often be the case for normal tissue. However, Suvà and Tirosh note in their recent review, “An emerging theme from recent scRNA-seq studies is that malignant cells tend to cluster in their expression profiles primarily by patient sample, and non-malignant cells cluster in their expression profiles by cell type, somewhat independently of the patient of origin (Filbin et al., 2018; Jerby-Arnon et al.,2018; Tirosh et al., 2016a, 2016b; Venteicher et al., 2017). This indicates that (1) inter-tumor heterogeneity is typically larger for malignant cells than for any particular type of non-malignant cells. (2) For malignant cells, inter-tumor heterogeneity is much larger than intra-tumor heterogeneity.” Imagine in the figure above that Cell Type 2’s expression differs greatly from patient-to-patient. There is some risk that deconvolution may be less successful if the inter-tumor expression heterogeneity for malignant cells is large for one or more of the profiled cancer types.
Why we asked for bulk profiling
On balance, there were enough unknown factors or areas of potential risk with a single-cell only approach for malignant tissue profiling that we elected to require bulk sequencing. As the RFA was being designed, it was not clear what the right approach would be for pediatric tumor tissues. These cancers are often somewhat biologically distinct from the adult cancers that much of the literature-to-date for single-cell profiling of human tumors has focused on. There have been a fair number of single-cell profiles of both PDX and mouse model samples, but it wasn’t clear if that data would be directly applicable to an atlas of human tumors.
Looking at the available literature and the uncertainties that existed, we thought it would be prudent to request candidates obtain bulk assays at the same time given the relative risks and benefits plus the modest cost of bulk sequencing relative to single-cell profiling. This means that we’ll be able to evaluate the concordance between single-cell profiles and what is observed in bulk samples. We hope that the references and literature can help others who are independently designing studies of malignancies, particularly pediatric ones, to best allocate resources towards their own efforts.

