Automating analyses with workflow managers
At the Data Lab, we are big proponents of automating the boring stuff so we can spend more time thinking about the fun stuff. But how exactly do we do that, and what does it mean to automate the boring stuff?
For many of the repetitive analysis and day-to-day tasks we perform, we implement workflows and automate these workflows with workflow managers. Workflows help us create single streamlined processes that can be run repeatedly for multiple samples and produce the same results. Not only do we save analysis time, but we also save time testing and adding features to our analysis. Changes only have to be made in one place, and that change is applied across all samples!
This approach allows us to spend less time repeating the same task over and over again and more time working on developing new analyses, exploring new datasets, and helping teach researchers like you!
So what are workflows and workflow managers?
A workflow refers to any number of steps or tasks joined together in a particular order to be executed in a repetitive manner and produce a desired outcome. Workflows can be as simple as a two-step process requiring one input to complicated multi-step processes taking multiple inputs and returning multiple outputs at various stages.
Workflow managers take these multi-part workflows and introduce automation of the individual tasks to increase productivity, eliminate manual repetition, and reduce errors. The ultimate goal of using a workflow manager is to translate a set of mundane tasks into a structured, automated set of tasks that can be repeated constantly with efficiency and accuracy to produce the desired output when provided a specific input.
When should you use workflow managers?
Building reproducible workflows run by workflow managers helps us by automating the repetitive, boring tasks, allowing us to clear our minds to focus on the more important tasks. However, knowing when it’s time to utilize automated workflows is a skill in itself. Setting up a workflow is a time intensive process, but when implemented in the right situations, it can save a lot of time in the long run.
What makes a task an ideal candidate for automation?
- Repetitive - If a task is something that is repeated more than three times, there’s a good chance that it would benefit from automation. When you start to copy and paste your code in multiple places, that’s a sign that automation could benefit you! Or if you need to analyze multiple samples through the same multi-step process, automation will save you time and manual effort.
- Error-prone - Tasks that are prone to human error (e.g. typos) are good candidates for workflow managers. Examples include having to manually enter in file paths throughout code or data entry.
Why do we need workflow managers and how do they help us?
Two examples of workflow managers that we use at the Data Lab are Nextflow and Snakemake. Both of these tools allow for multi-step workflow management, and creating workflows that are reproducible and scalable. They both do this by utilizing modular workflows. A modular workflow is one that separates each individual task into its own unit of work with each process having its own input and output inside of the overarching workflow. Using a modular setup, like those implemented with Nextflow and Snakemake, allows you to easily add new features and test and debug your workflow.
Below are some more key ways that these tools improve our workflows:
- Running multiple steps in sequence, or in parallel when possible
- Skipping steps that have already been completed, allowing users to easily modify later steps and resume the workflow from the last successfully completed step
- Managing resources needed for each step of the workflow
- Keeping a log of successes and failures
- Using containers, allowing all dependencies to be packaged together and increasing reproducibility
- Allowing for the same workflow to be run on multiple platforms - locally, in the cloud, or on a server
Each of these tools have their own unique benefits, and we encourage you to read more on how they manage workflows.
Example: Automating an RNA-Seq Analysis Workflow
We previously described the processes behind setting up a workflow for RNA-sequencing analysis, so we’ll build on that example here. In this scenario, you are starting with a set of FASTQ files for a large amount of RNA-sequencing samples that your PI has asked you to analyze. They have asked you to identify the expression of a set of marker genes that you are studying. You’ve already identified that you have to perform three separate tasks to analyze these files: trim the reads, align and quantify, and plot the expression of your marker genes.
One option is to analyze each of these samples individually, setting up the code each time to take in a different FASTQ file. However, that can be time-consuming and error-prone. Imagine having to make a change and implementing it for every single sample, that would take forever and be likely to cause errors. With automation, we only have to make any future changes one time! Additionally, with a workflow manager, we can process multiple samples in parallel, saving us overall analysis time. This is the perfect situation in which automation can help us, so let’s set up a workflow and use a workflow manager!
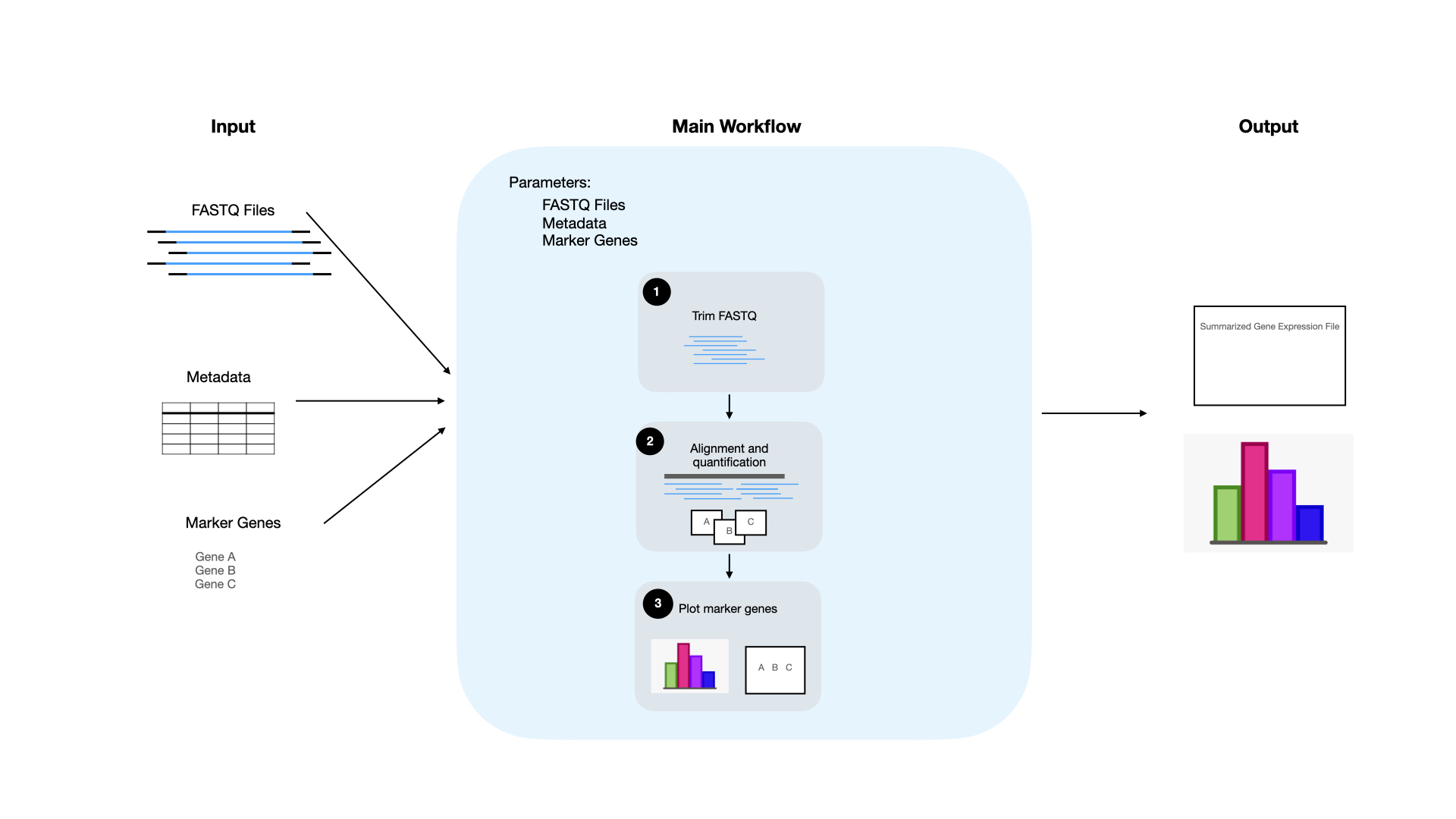
- Outline the workflow - The first thing we will want to do is outline our workflow. This entails determining what we want to have as our input to the workflow and what the workflow will return as output. Our workflow will take FASTQ files as input and then output a summarized file with the gene expression of all samples and a plot showing the marker gene expression. Then we decide on the individual steps that we will take to get from the input (FASTQ files) to the output (plots showing marker gene expression). For our specific example, the steps that we will perform are trimming the FASTQ files, aligning the FASTQ files, and then plotting our results. Each of these steps will also have an input and then an output and the outline will help us reveal how we plan to connect the outputs of step 1 (trimming) to the inputs of step 2 (aligning).
- Define the parameters - Now that we know what inputs our workflow will require, we can design a set of parameters. These parameters will set the default settings and input needed to run the workflow. For our workflow, the parameters we will set are the location for the FASTQ files, the metadata information about each sample, and a list of marker genes that we are interested in. To ensure that our workflow is reproducible, we don’t want to hard code this information into the workflow. Instead we want to set each of these inputs as a parameter so we can easily re-use this workflow for a different set of samples in the future.
- Develop individual steps - Next, we are ready to start adding the individual processes to the workflow. Each process will correspond to one step of the workflow, and steps will be executed in the order that you specify, parallelizing when possible. See the schematic below to see how the individual steps of the workflow will work together to produce the desired output.
- Test your workflow - Before implementing our workflow, we want to make sure that we test it on 1-2 of our samples, rather than running it on all of them. During testing we don’t want to run on all of them in case we do run into errors. Running our pipeline on all of our samples before it’s ready could use up valuable resources, like computing time and money. If we encounter any errors during testing, we can make adjustments to the processes that failed and re-run. The benefit of using a workflow manager is the ability to intelligently restart the pipeline after any step that has completed successfully. This means that we can make adjustments to any downstream steps that had errors and re-run our pipeline without having to re-run the entire workflow, making the testing and debugging process smoother.
- Add features - If there are additional steps we want to incorporate, such as additional downstream analyses, we can easily add those in by creating another process in the main workflow.
Now you’re ready to give it a try and automate some of your own workflows! If you’re interested in learning more about the tools we use and the processes we implement at the Data Lab, fill out this form to be notified about future materials and offerings like this.
At the Data Lab, we are big proponents of automating the boring stuff so we can spend more time thinking about the fun stuff. But how exactly do we do that, and what does it mean to automate the boring stuff?
For many of the repetitive analysis and day-to-day tasks we perform, we implement workflows and automate these workflows with workflow managers. Workflows help us create single streamlined processes that can be run repeatedly for multiple samples and produce the same results. Not only do we save analysis time, but we also save time testing and adding features to our analysis. Changes only have to be made in one place, and that change is applied across all samples!
This approach allows us to spend less time repeating the same task over and over again and more time working on developing new analyses, exploring new datasets, and helping teach researchers like you!
So what are workflows and workflow managers?
A workflow refers to any number of steps or tasks joined together in a particular order to be executed in a repetitive manner and produce a desired outcome. Workflows can be as simple as a two-step process requiring one input to complicated multi-step processes taking multiple inputs and returning multiple outputs at various stages.
Workflow managers take these multi-part workflows and introduce automation of the individual tasks to increase productivity, eliminate manual repetition, and reduce errors. The ultimate goal of using a workflow manager is to translate a set of mundane tasks into a structured, automated set of tasks that can be repeated constantly with efficiency and accuracy to produce the desired output when provided a specific input.
When should you use workflow managers?
Building reproducible workflows run by workflow managers helps us by automating the repetitive, boring tasks, allowing us to clear our minds to focus on the more important tasks. However, knowing when it’s time to utilize automated workflows is a skill in itself. Setting up a workflow is a time intensive process, but when implemented in the right situations, it can save a lot of time in the long run.
What makes a task an ideal candidate for automation?
- Repetitive - If a task is something that is repeated more than three times, there’s a good chance that it would benefit from automation. When you start to copy and paste your code in multiple places, that’s a sign that automation could benefit you! Or if you need to analyze multiple samples through the same multi-step process, automation will save you time and manual effort.
- Error-prone - Tasks that are prone to human error (e.g. typos) are good candidates for workflow managers. Examples include having to manually enter in file paths throughout code or data entry.
Why do we need workflow managers and how do they help us?
Two examples of workflow managers that we use at the Data Lab are Nextflow and Snakemake. Both of these tools allow for multi-step workflow management, and creating workflows that are reproducible and scalable. They both do this by utilizing modular workflows. A modular workflow is one that separates each individual task into its own unit of work with each process having its own input and output inside of the overarching workflow. Using a modular setup, like those implemented with Nextflow and Snakemake, allows you to easily add new features and test and debug your workflow.
Below are some more key ways that these tools improve our workflows:
- Running multiple steps in sequence, or in parallel when possible
- Skipping steps that have already been completed, allowing users to easily modify later steps and resume the workflow from the last successfully completed step
- Managing resources needed for each step of the workflow
- Keeping a log of successes and failures
- Using containers, allowing all dependencies to be packaged together and increasing reproducibility
- Allowing for the same workflow to be run on multiple platforms - locally, in the cloud, or on a server
Each of these tools have their own unique benefits, and we encourage you to read more on how they manage workflows.
Example: Automating an RNA-Seq Analysis Workflow
We previously described the processes behind setting up a workflow for RNA-sequencing analysis, so we’ll build on that example here. In this scenario, you are starting with a set of FASTQ files for a large amount of RNA-sequencing samples that your PI has asked you to analyze. They have asked you to identify the expression of a set of marker genes that you are studying. You’ve already identified that you have to perform three separate tasks to analyze these files: trim the reads, align and quantify, and plot the expression of your marker genes.
One option is to analyze each of these samples individually, setting up the code each time to take in a different FASTQ file. However, that can be time-consuming and error-prone. Imagine having to make a change and implementing it for every single sample, that would take forever and be likely to cause errors. With automation, we only have to make any future changes one time! Additionally, with a workflow manager, we can process multiple samples in parallel, saving us overall analysis time. This is the perfect situation in which automation can help us, so let’s set up a workflow and use a workflow manager!
- Outline the workflow - The first thing we will want to do is outline our workflow. This entails determining what we want to have as our input to the workflow and what the workflow will return as output. Our workflow will take FASTQ files as input and then output a summarized file with the gene expression of all samples and a plot showing the marker gene expression. Then we decide on the individual steps that we will take to get from the input (FASTQ files) to the output (plots showing marker gene expression). For our specific example, the steps that we will perform are trimming the FASTQ files, aligning the FASTQ files, and then plotting our results. Each of these steps will also have an input and then an output and the outline will help us reveal how we plan to connect the outputs of step 1 (trimming) to the inputs of step 2 (aligning).
- Define the parameters - Now that we know what inputs our workflow will require, we can design a set of parameters. These parameters will set the default settings and input needed to run the workflow. For our workflow, the parameters we will set are the location for the FASTQ files, the metadata information about each sample, and a list of marker genes that we are interested in. To ensure that our workflow is reproducible, we don’t want to hard code this information into the workflow. Instead we want to set each of these inputs as a parameter so we can easily re-use this workflow for a different set of samples in the future.
- Develop individual steps - Next, we are ready to start adding the individual processes to the workflow. Each process will correspond to one step of the workflow, and steps will be executed in the order that you specify, parallelizing when possible. See the schematic below to see how the individual steps of the workflow will work together to produce the desired output.
- Test your workflow - Before implementing our workflow, we want to make sure that we test it on 1-2 of our samples, rather than running it on all of them. During testing we don’t want to run on all of them in case we do run into errors. Running our pipeline on all of our samples before it’s ready could use up valuable resources, like computing time and money. If we encounter any errors during testing, we can make adjustments to the processes that failed and re-run. The benefit of using a workflow manager is the ability to intelligently restart the pipeline after any step that has completed successfully. This means that we can make adjustments to any downstream steps that had errors and re-run our pipeline without having to re-run the entire workflow, making the testing and debugging process smoother.
- Add features - If there are additional steps we want to incorporate, such as additional downstream analyses, we can easily add those in by creating another process in the main workflow.

Now you’re ready to give it a try and automate some of your own workflows! If you’re interested in learning more about the tools we use and the processes we implement at the Data Lab, fill out this form to be notified about future materials and offerings like this.

