Gene Expression Repositories Explained
The goal of our refine.bio project is to download, process, and make available gene expression datasets that can be analyzed together, or in parts, depending on a researcher’s need. Childhood cancer researchers need to be able to use data generated through multiple profiling technologies including microarrays and RNA-sequencing. A big part of what refine.bio does is downloading gene expression data from various repositories. We wanted to download and provide ALL THE DATA, but certain repositories mirror some fraction of other repositories. To be able to do this successfully without producing duplicate data, we had to learn a lot about the various repositories for gene expression data and their data models. We also needed to determine if and how the various repositories overlapped. This post has two main sections:
- Repositories and Organizations: The different repositories we download from, the organizations that run them, and how they interconnect and overlap.
- The SRA Data Model: An explanation of the data model used by the Sequence Read Archive (SRA).
The Repositories and Organizations
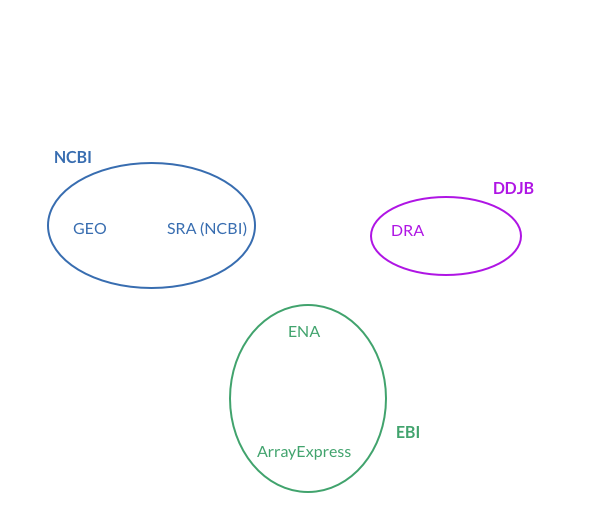
The data refine.bio has downloaded, processed, and is now serving can be found across five repositories (Array Express, GEO, SRA, ENA, and DRA) which are run by three different organizations (NCBI, EMBL-EBI, and DDBJ). That’s a whole alphabet of acronyms so why don’t we start by explaining what/who they all are.
The National Center for Biotechnology Information (NCBI)
NCBI is a part of the U.S. National Library of Medicine which is part of The National Institutes of Health (NIH). NCBI has a lot of resources, tools, programs, and data repositories but the two that store gene expression data are The Gene Expression Omnibus (GEO) and the Sequence Read Archive (SRA). Microarray data are stored in GEO. RNA-seq data are primarily stored in SRA; however, certain RNA-seq datasets without raw data available appear to be provided via GEO.
The European Molecular Biology Laboratory’s European Bioinformatic’s Institute (EMBL-EBI or EBI)
EBI is another large organization which also offers a lot of resources, tools, programs, and data repositories, but the two that store gene expression data are Array Express and The European Nucleotide Archive (ENA). Array Express and the ENA have a similar relationship to GEO & SRA at NCBI, though they serve as the primary data deposit location for investigators in Europe.
The DNA Data Bank of Japan (DDBJ)
The DDBJ seems to be smaller than the NCBI or EMBL-EBI in terms of data uniquely found at the DDBJ, but it does include the DDBJ Sequence Read Archive (DRA) which is relevant for this post.
The Sequence Read Archive

Over 30 years ago subsections of NCBI, EMBL-EBI, and DDBJ came together to form the The International Nucleotide Sequence Database Collaboration (INSDC). (source: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3013722/) The INSDC joined NCBI’s Genbank, EMB-EBI’s ENA, and DDBJ’s DRA together into a unified group “to ensure that all public domain nucleotide sequence data deposited in the archives is preserved as part of the scientific record and is accessible in standardized formats across the three sites through daily data exchange.” (source: https://academic.oup.com/nar/article/46/D1/D48/4668651). The INSDC has two primary offerings: “Raw data archives under the collaboration are known as the Trace Archive for raw data from capillary electrophoresis platforms and the Sequence Read Archive for raw and read alignment data from next-generation platforms.” (source: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3013722/).
You may have noticed that both NCBI and INSDC both have an offering called SRA and you may be wondering if these are the same thing. As far as I can tell, the answer is both yes and no. The INSDC’s SRA is a special database that is co-managed by NCBI, EBI, and DDBJ. Each of those organizations host a full copy of the database. EBI’s copy of the database is called ENA, DDBJ’s copy of the database is called DRA, and NCBI’s copy of the database is confusingly called SRA. Each of these organizations also can accept new submissions to the database.
If an experiment is first submitted to ENA, it and associated data objects will be prefixed with `ER` such as ERP008771, ERX1762259, or ERR1692631. If an experiment is first submitted to DRA, it and associated data objects will instead be prefixed with `DR` such as DRP000425, DRX000772, or DRR001175. If an experiment is first submitted to NCBI’s SRA, it and associated data objects will be prefixed with `SR` such as SRP060416, SRX1082691, or SRR2088722. If you’re asking what are the differences between ERP/DRP/SRP, ERX/DRX/SRX, and ERR/DRR/SRR data objects, then stay tuned. I’ll cover SRA’s data model in the next section.
Fun Fact: According to the SRA wikipedia page it used to stand for Short Read Archive, so if you ever see that it’s not entirely wrong.
Bonus: While researching this post, I came across China’s Genome Sequence Archive (GSA) which adheres “with data standards and structures of the INSDC” (source: https://www.sciencedirect.com/science/article/pii/S1672022917300025). However they do not appear to be replicating and contributing to the shared SRA repository and instead are maintaining their own collection. Downloading, processing, and serving data from GSA has already been added to our future plans, especially given the growth in Chinese investment in basic science.
Microarray Repositories

refine.bio harmonizes two different types of gene expression data: RNA-seq and microarray. SRA is where refine.bio downloads all its RNA-seq data. refine.bio downloads microarray data from both NCBI’s GEO and EMBL-EBI’s Array Express. These two repositories also have a somewhat convoluted relationship. Array Express used to replicate data from GEO on a weekly basis, so it contains a lot of data from there. This was true when we first started the refine.bio project, so our original idea was to only download microarray data from Array Express. However since then they have stopped replicating data, so we now download data from the source (Array Express / GEO) that it was originally uploaded to, which we determine by its identifier.
Fun Fact: Any accession code on Array Express that starts with “E-GEOD-” was duplicated from GEO. If you remove the leading “E-GEOD-” and replace it with “GSE”, you get the accession code used by GEO! For example, E-GEOD-7307 is the same experiment as GSE7307.
GEO is an NCBI offering. It does not include raw RNA-seq data that can be retrieved from SRA. However, there are some experiments that mix RNA-seq and microarray data in the GEO database. These will sometimes have the raw microarray data associated with them, but only metadata or processed data for RNA-seq data. However, using the metadata it is possible to find the same sample in SRA to get the raw data. As far as we have seen so far, Array Express does not host any RNA-seq data.
The Full Picture
If you have gotten confused trying to follow along as I explained the relationships between INSDC, NCBI, EMBL-EBI, DDBJ, and their various sub-organizations and offerings, we’ve put together this diagram which may help:

The SRA Data Model
Each these organizations do a lot more than just host the Gene Expression data we need for refine.bio, but that is outside the scope of this post. However, I promised to explain the SRA data model. To be clear, for the rest of this post when I say “SRA” I am referring to INSDC’s SRA instead of NCBI’s SRA. Everything will apply to both, but every member of the INSDC uses the same data model for their SRA databases.
Let’s start with the six types of SRA metadata objects, then we can get into how they relate to each other:
Run - A metadata object that directly represents the file generated by sequencing. Accessions for Runs have the prefix SRR/ERR/DRR.
Experiment - Metadata about how the sequencing was performed. Accessions for Experiments have the prefix SRX/ERX/DRX.
Sample - A description of biologically or physically unique specimens. Directly corresponds to a BioSample. Accessions for Samples have the prefix SRS/ERS/DRS.
Study - A description of the research effort that required the sequencing. Directly corresponds to a BioProject. Accessions for Studies have the prefix SRP/ERP/DRP.
Submission - Metadata about the submission of the data to SRA. Accessions for Submissions have the prefix SRA/ERA/DRA.
Analysis - A representation of an analysis that was submitted to SRA about the data. refine.bio does not survey or store metadata objects of this type.
(source: https://www.ddbj.nig.ac.jp/dra/submission-e.html)
The simplest image we’ve seen of the SRA data model comes from the ENA’s page on programmatically submitting data, although unfortunately it leaves out Submissions:

The description on that page is also one of the better descriptions of how the different objects relate to each other. The key takeaways for understanding this data model are:
- A single Sample can be used by more than one Experiment, so there is a one-to-many relationship between Samples and Experiments.
- Multiple Experiments can be grouped together to form a Study, so there is also a one-to-many relationship between Studies and Experiments.
- One Experiment can be run through the sequencing apparatus multiple time to generate multiple Runs, so there is a one-to-many relationship between Experiments and Runs.
- A Run has a one-to-one relationship with actual files containing reads, unless the Run was Paired-End, in which case there will be two files (assuming the FASTQ format was used).
The one metadata object type that is omitted here is Submission. The documentation is a bit less clear about the links for Submissions, most likely because they are generally linked up automatically during the submission process, which is what that documentation is about. However, Submission objects aren’t that involved. They are actually representing the submission of the data to SRA and therefore they can be linked to any of the other objects. They don’t have too much information other than linking everything together explicitly.
Simplifying for the Future
This pretty much covers what we know about gene expression repositories that wasn’t very easy for us to find and learn while developing refine.bio. We hope that as refine.bio discovers, downloads, processes, and makes available more and more datasets that fewer and fewer people will need to understand these arcane secrets. However, we think it’s important that people are able to use these repositories directly and we hope this post makes that a bit easier.
The goal of our refine.bio project is to download, process, and make available gene expression datasets that can be analyzed together, or in parts, depending on a researcher’s need. Childhood cancer researchers need to be able to use data generated through multiple profiling technologies including microarrays and RNA-sequencing. A big part of what refine.bio does is downloading gene expression data from various repositories. We wanted to download and provide ALL THE DATA, but certain repositories mirror some fraction of other repositories. To be able to do this successfully without producing duplicate data, we had to learn a lot about the various repositories for gene expression data and their data models. We also needed to determine if and how the various repositories overlapped. This post has two main sections:
- Repositories and Organizations: The different repositories we download from, the organizations that run them, and how they interconnect and overlap.
- The SRA Data Model: An explanation of the data model used by the Sequence Read Archive (SRA).
The Repositories and Organizations
The data refine.bio has downloaded, processed, and is now serving can be found across five repositories (Array Express, GEO, SRA, ENA, and DRA) which are run by three different organizations (NCBI, EMBL-EBI, and DDBJ). That’s a whole alphabet of acronyms so why don’t we start by explaining what/who they all are.

The National Center for Biotechnology Information (NCBI)
NCBI is a part of the U.S. National Library of Medicine which is part of The National Institutes of Health (NIH). NCBI has a lot of resources, tools, programs, and data repositories but the two that store gene expression data are The Gene Expression Omnibus (GEO) and the Sequence Read Archive (SRA). Microarray data are stored in GEO. RNA-seq data are primarily stored in SRA; however, certain RNA-seq datasets without raw data available appear to be provided via GEO.
The European Molecular Biology Laboratory’s European Bioinformatic’s Institute (EMBL-EBI or EBI)
EBI is another large organization which also offers a lot of resources, tools, programs, and data repositories, but the two that store gene expression data are Array Express and The European Nucleotide Archive (ENA). Array Express and the ENA have a similar relationship to GEO & SRA at NCBI, though they serve as the primary data deposit location for investigators in Europe.
The DNA Data Bank of Japan (DDBJ)
The DDBJ seems to be smaller than the NCBI or EMBL-EBI in terms of data uniquely found at the DDBJ, but it does include the DDBJ Sequence Read Archive (DRA) which is relevant for this post.
The Sequence Read Archive

Over 30 years ago subsections of NCBI, EMBL-EBI, and DDBJ came together to form the The International Nucleotide Sequence Database Collaboration (INSDC). (source: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3013722/) The INSDC joined NCBI’s Genbank, EMB-EBI’s ENA, and DDBJ’s DRA together into a unified group “to ensure that all public domain nucleotide sequence data deposited in the archives is preserved as part of the scientific record and is accessible in standardized formats across the three sites through daily data exchange.” (source: https://academic.oup.com/nar/article/46/D1/D48/4668651). The INSDC has two primary offerings: “Raw data archives under the collaboration are known as the Trace Archive for raw data from capillary electrophoresis platforms and the Sequence Read Archive for raw and read alignment data from next-generation platforms.” (source: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3013722/).
You may have noticed that both NCBI and INSDC both have an offering called SRA and you may be wondering if these are the same thing. As far as I can tell, the answer is both yes and no. The INSDC’s SRA is a special database that is co-managed by NCBI, EBI, and DDBJ. Each of those organizations host a full copy of the database. EBI’s copy of the database is called ENA, DDBJ’s copy of the database is called DRA, and NCBI’s copy of the database is confusingly called SRA. Each of these organizations also can accept new submissions to the database.
If an experiment is first submitted to ENA, it and associated data objects will be prefixed with `ER` such as ERP008771, ERX1762259, or ERR1692631. If an experiment is first submitted to DRA, it and associated data objects will instead be prefixed with `DR` such as DRP000425, DRX000772, or DRR001175. If an experiment is first submitted to NCBI’s SRA, it and associated data objects will be prefixed with `SR` such as SRP060416, SRX1082691, or SRR2088722. If you’re asking what are the differences between ERP/DRP/SRP, ERX/DRX/SRX, and ERR/DRR/SRR data objects, then stay tuned. I’ll cover SRA’s data model in the next section.
Fun Fact: According to the SRA wikipedia page it used to stand for Short Read Archive, so if you ever see that it’s not entirely wrong.
Bonus: While researching this post, I came across China’s Genome Sequence Archive (GSA) which adheres “with data standards and structures of the INSDC” (source: https://www.sciencedirect.com/science/article/pii/S1672022917300025). However they do not appear to be replicating and contributing to the shared SRA repository and instead are maintaining their own collection. Downloading, processing, and serving data from GSA has already been added to our future plans, especially given the growth in Chinese investment in basic science.
Microarray Repositories

refine.bio harmonizes two different types of gene expression data: RNA-seq and microarray. SRA is where refine.bio downloads all its RNA-seq data. refine.bio downloads microarray data from both NCBI’s GEO and EMBL-EBI’s Array Express. These two repositories also have a somewhat convoluted relationship. Array Express used to replicate data from GEO on a weekly basis, so it contains a lot of data from there. This was true when we first started the refine.bio project, so our original idea was to only download microarray data from Array Express. However since then they have stopped replicating data, so we now download data from the source (Array Express / GEO) that it was originally uploaded to, which we determine by its identifier.
Fun Fact: Any accession code on Array Express that starts with “E-GEOD-” was duplicated from GEO. If you remove the leading “E-GEOD-” and replace it with “GSE”, you get the accession code used by GEO! For example, E-GEOD-7307 is the same experiment as GSE7307.
GEO is an NCBI offering. It does not include raw RNA-seq data that can be retrieved from SRA. However, there are some experiments that mix RNA-seq and microarray data in the GEO database. These will sometimes have the raw microarray data associated with them, but only metadata or processed data for RNA-seq data. However, using the metadata it is possible to find the same sample in SRA to get the raw data. As far as we have seen so far, Array Express does not host any RNA-seq data.
The Full Picture
If you have gotten confused trying to follow along as I explained the relationships between INSDC, NCBI, EMBL-EBI, DDBJ, and their various sub-organizations and offerings, we’ve put together this diagram which may help:

The SRA Data Model
Each these organizations do a lot more than just host the Gene Expression data we need for refine.bio, but that is outside the scope of this post. However, I promised to explain the SRA data model. To be clear, for the rest of this post when I say “SRA” I am referring to INSDC’s SRA instead of NCBI’s SRA. Everything will apply to both, but every member of the INSDC uses the same data model for their SRA databases.
Let’s start with the six types of SRA metadata objects, then we can get into how they relate to each other:
Run - A metadata object that directly represents the file generated by sequencing. Accessions for Runs have the prefix SRR/ERR/DRR.
Experiment - Metadata about how the sequencing was performed. Accessions for Experiments have the prefix SRX/ERX/DRX.
Sample - A description of biologically or physically unique specimens. Directly corresponds to a BioSample. Accessions for Samples have the prefix SRS/ERS/DRS.
Study - A description of the research effort that required the sequencing. Directly corresponds to a BioProject. Accessions for Studies have the prefix SRP/ERP/DRP.
Submission - Metadata about the submission of the data to SRA. Accessions for Submissions have the prefix SRA/ERA/DRA.
Analysis - A representation of an analysis that was submitted to SRA about the data. refine.bio does not survey or store metadata objects of this type.
(source: https://www.ddbj.nig.ac.jp/dra/submission-e.html)
The simplest image we’ve seen of the SRA data model comes from the ENA’s page on programmatically submitting data, although unfortunately it leaves out Submissions:

The description on that page is also one of the better descriptions of how the different objects relate to each other. The key takeaways for understanding this data model are:
- A single Sample can be used by more than one Experiment, so there is a one-to-many relationship between Samples and Experiments.
- Multiple Experiments can be grouped together to form a Study, so there is also a one-to-many relationship between Studies and Experiments.
- One Experiment can be run through the sequencing apparatus multiple time to generate multiple Runs, so there is a one-to-many relationship between Experiments and Runs.
- A Run has a one-to-one relationship with actual files containing reads, unless the Run was Paired-End, in which case there will be two files (assuming the FASTQ format was used).
The one metadata object type that is omitted here is Submission. The documentation is a bit less clear about the links for Submissions, most likely because they are generally linked up automatically during the submission process, which is what that documentation is about. However, Submission objects aren’t that involved. They are actually representing the submission of the data to SRA and therefore they can be linked to any of the other objects. They don’t have too much information other than linking everything together explicitly.
Simplifying for the Future
This pretty much covers what we know about gene expression repositories that wasn’t very easy for us to find and learn while developing refine.bio. We hope that as refine.bio discovers, downloads, processes, and makes available more and more datasets that fewer and fewer people will need to understand these arcane secrets. However, we think it’s important that people are able to use these repositories directly and we hope this post makes that a bit easier.

